Data Encoding for Quantum Pangenomics
Tuesday, June 10, 2025 3:00 PM to Thursday, June 12, 2025 4:00 PM · 2 days 1 hr. (Europe/Berlin)
Foyer D-G - 2nd floor
Research Poster
Bioinformatics and Life SciencesIntegration of Quantum Computing and HPCQuantum Computing - Use CasesSimulating Quantum Systems
Information
Poster is on display and will be presented at the poster pitch session.
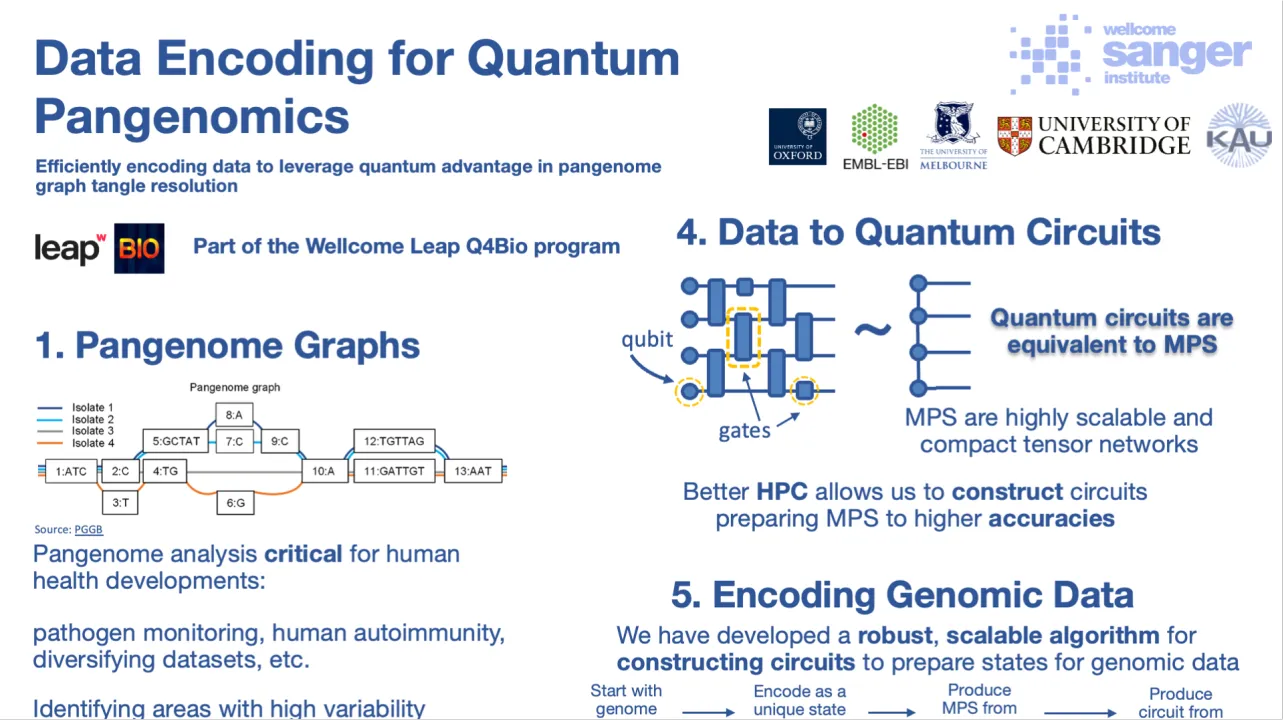
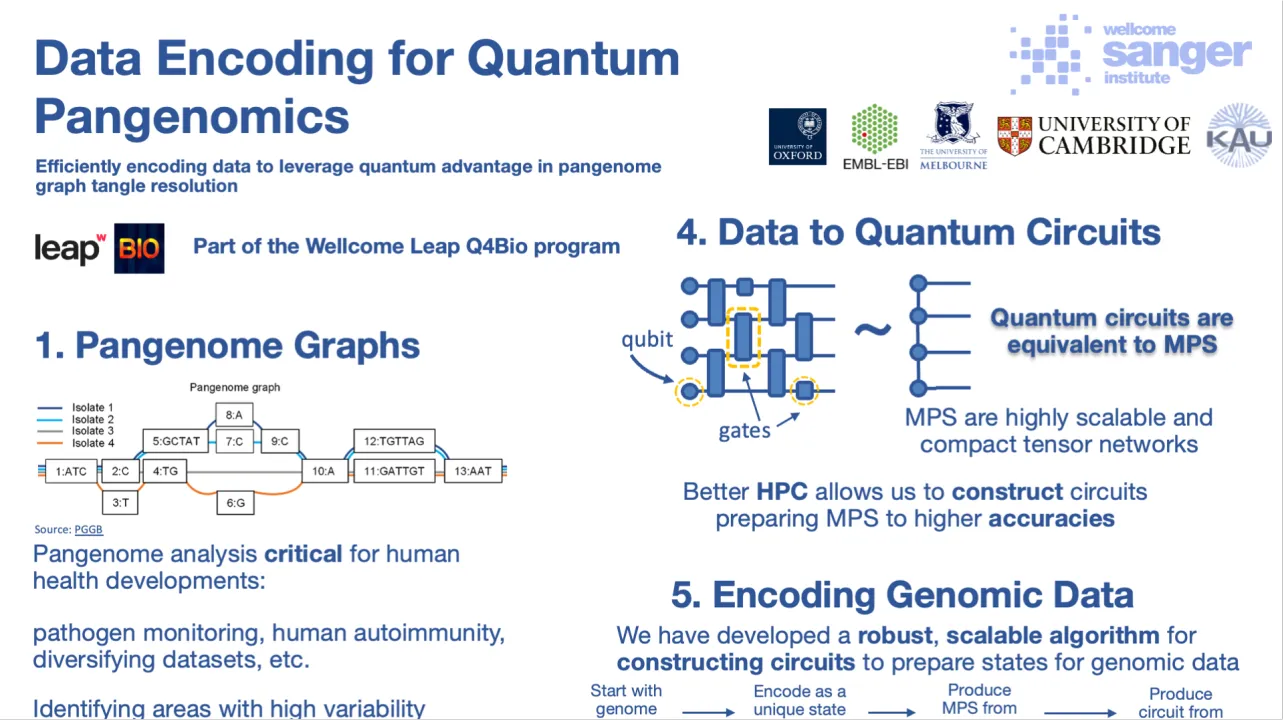
Recent advances in genomic research, driven by the rapid growth of sequencing technologies and data analysis methods, have raised questions about the adequacy of single reference genomes in capturing the full genetic diversity of species. Pangenomics, the study of multiple complete genomes in tandem, provides a more comprehensive approach by analysing the entire genetic variation within a species, rather than relying on a single reference genome.
However, pangenomic analysis is computationally intensive due to the complexity and structure of the data. Classical algorithms often depend on heuristics to manage these complexities, which limits their scalability and accuracy as datasets grow.
Quantum computing offers promise to revolutionise the field, by using algorithms capable of efficiently navigating complex data. This quantum advantage could enable more accurate and scalable pangenome analysis without requiring heuristics, facilitating insights into regions with high genetic variability (e.g., the HLA-DRB1 gene, critical for human immune function) and improving pathogen surveillance (e.g., tracking mutations in the spike protein of SARS-CoV-2).
As part of the Wellcome Leap Q4Bio initiative, our international team is pioneering the application of quantum computing to pangenomics, and laying the foundations of a novel field of research in Quantum Pangenomics, and demonstrating the power of this research to improve human health on a global scale.
Crucial to this effort are efficient methods for encoding genomic data for quantum systems. We demonstrate a novel encoding utilising tensor networks, specifically matrix product states (MPS), to represent genome sequences such that similarities between sequences are reflected in the fidelities between their respective encodings. Our method ensures that genomic data can be accurately mapped to quantum states, with the well-understood correspondence between MPS and quantum circuits allowing us to efficiently produce corresponding circuits.
We demonstrate the scalability and efficiency of our encoding through simulations on HPC. We successfully encode the genome of the bacteriophage Phi-X-174 (length: 5386 base pairs) into just 15 qubits, requiring O(1,000) gates. Our MPS-based encoding significantly outperforms standard simulation methods in both gate count and runtime. This simulation method can also be applied today to useful genes across species, including the SARS-CoV-2 S-gene and HLA-DRB1, offering a powerful tool for advancing pangenomic analysis.
In the near future, we will drastically upscale our HPC simulations, and implement our circuits on quantum hardware. Together with upcoming advances in quantum computing, we are confident of our ability to utilise this research in improving human health across the globe.
Contributors:
Recent advances in genomic research, driven by the rapid growth of sequencing technologies and data analysis methods, have raised questions about the adequacy of single reference genomes in capturing the full genetic diversity of species. Pangenomics, the study of multiple complete genomes in tandem, provides a more comprehensive approach by analysing the entire genetic variation within a species, rather than relying on a single reference genome.
However, pangenomic analysis is computationally intensive due to the complexity and structure of the data. Classical algorithms often depend on heuristics to manage these complexities, which limits their scalability and accuracy as datasets grow.
Quantum computing offers promise to revolutionise the field, by using algorithms capable of efficiently navigating complex data. This quantum advantage could enable more accurate and scalable pangenome analysis without requiring heuristics, facilitating insights into regions with high genetic variability (e.g., the HLA-DRB1 gene, critical for human immune function) and improving pathogen surveillance (e.g., tracking mutations in the spike protein of SARS-CoV-2).
As part of the Wellcome Leap Q4Bio initiative, our international team is pioneering the application of quantum computing to pangenomics, and laying the foundations of a novel field of research in Quantum Pangenomics, and demonstrating the power of this research to improve human health on a global scale.
Crucial to this effort are efficient methods for encoding genomic data for quantum systems. We demonstrate a novel encoding utilising tensor networks, specifically matrix product states (MPS), to represent genome sequences such that similarities between sequences are reflected in the fidelities between their respective encodings. Our method ensures that genomic data can be accurately mapped to quantum states, with the well-understood correspondence between MPS and quantum circuits allowing us to efficiently produce corresponding circuits.
We demonstrate the scalability and efficiency of our encoding through simulations on HPC. We successfully encode the genome of the bacteriophage Phi-X-174 (length: 5386 base pairs) into just 15 qubits, requiring O(1,000) gates. Our MPS-based encoding significantly outperforms standard simulation methods in both gate count and runtime. This simulation method can also be applied today to useful genes across species, including the SARS-CoV-2 S-gene and HLA-DRB1, offering a powerful tool for advancing pangenomic analysis.
In the near future, we will drastically upscale our HPC simulations, and implement our circuits on quantum hardware. Together with upcoming advances in quantum computing, we are confident of our ability to utilise this research in improving human health across the globe.
Contributors:
Format
On DemandOn Site

